Tutorial#
Here we’ll walk through a typical run of ggCaller, including both Gene-calling and Querying.
Example results can be found here.
Important
Results will be consistent, but may not exactly match between your run and the example. This is due to the greedy clustering algorithm used by ggCaller, which can cause small differences in genes counts.
Installation and setup#
Follow the guide in Installation for downloading and installing ggCaller.
Working Dataset#
We’ll use a dataset from Bentley et al. (2006). This dataset contains 91 sequences pneumococcal capsular polysaccharide synthetic (CPS) loci. These sequences are structurally diverse, but are only ~20,000 bp in length, so can be analysed quickly (~5-10 minutes) on a standard laptop or desktop.
Download the files from here and unzip:
tar xvf Bentley_et_al_2006_CPS_sequences.tar.bz2
We will also provide our own custom annotation database for DIAMOND. These will be the manually curated protein sequences from Bentley et al. Download from here and unzip:
tar xvf Bentley_et_al_2006_CPS_protein_sequences.tar.bz2
Gene-calling#
First generate an input file for ggCaller. This must be a file containing paths (absolute recommended) to all sequences to be analysed. We recommend running the below command within the unzipped to generate this file:
cd Bentley_et_al_2006_CPS_sequences
ls -d -1 $PWD/*.fa > input.txt
cd ..
input.txt will now contain absolute paths to all .fa files in the directory Bentley_et_al_2006_CPS_sequences.
Now we will run ggCaller specifying the below settings:
Sensitive DIAMOND annotation using a custom database, and HMMER3 using the default database
Pangenome-wide alignment using default MAFFT
Saved intermediate datastructures, enabling sequence querying
To do this using 4 threads, run:
ggcaller --refs Bentley_et_al_2006_CPS_sequences/input.txt --annotation ultrasensitive --diamonddb Bentley_et_al_2006_CPS_protein_sequences.faa --aligner def --alignment pan --save --out ggc_Bentley_et_al_CPS --threads 4
You will find the following files in the output directory ggc_Bentley_et_al_CPS:
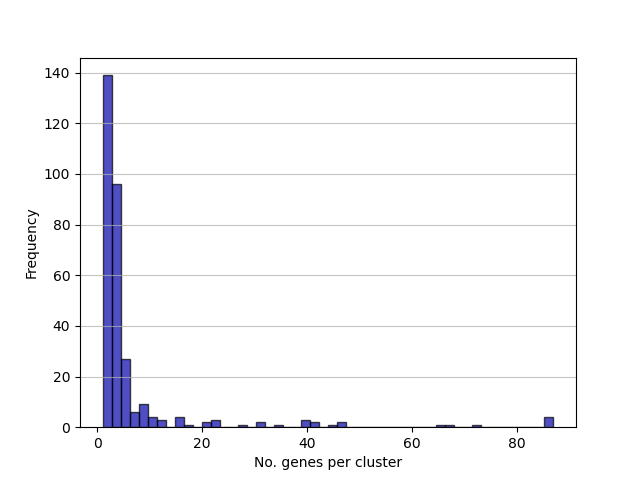
cluster_size.png: a frequency distribution of clusters by the number of genes found within them
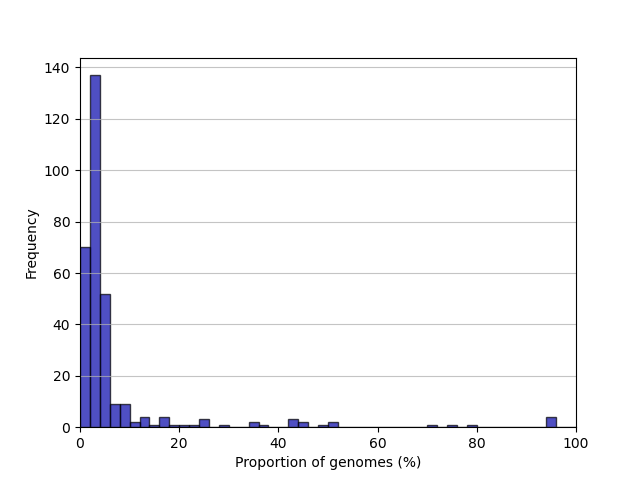
gene_frequency.png: a frequency distribution of clusters by proportion of dataset
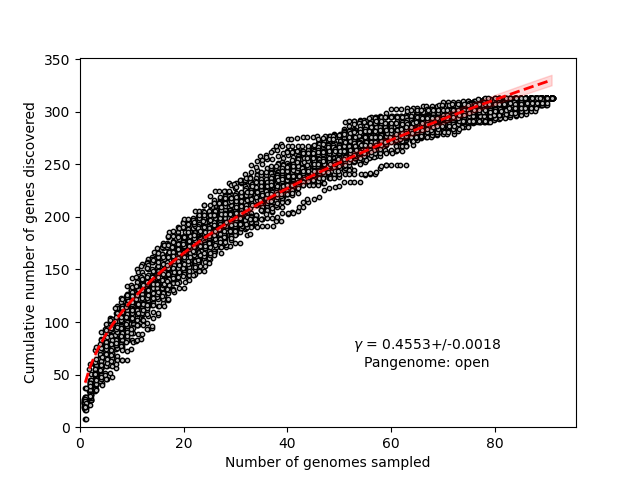
rarefaction_curve.png: rarefaction curve, describes the number of new genes discovered with random addition of a single genome. Also includes power-law fit for determination of pangenome openness, based on Tettelin et al. (2005).
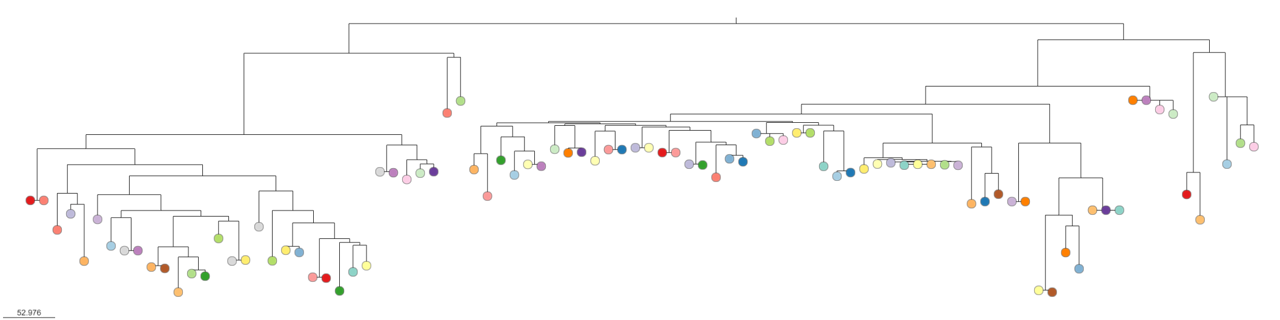
core_gene_alignment.aln: concatenated core genome alignmentcore_alignment_header.embl: core genome alignment in EMBL formatcore_tree_NJ.nwk: Neighbour joining tree from core genome alignment generated by RapidNJ. This can be visualised in Microreact
pangenome_NJ.nwk: Neighbour joining tree from gene presence/absence matrix generated by RapidNJ (can also be visualised in Microreact).
pan_genome_reference.fa: contains centroids for each cluster in FASTA formatgene_calls.faaandgene_calls.ffn: gene predictions with annotations in amino-acid and nucleotide FASTA formatspre_filt_graph.gmlandfinal_graph.gml: gene graphs pre- and post-quality control with Panaroogene_presence_absence*: gene presence absence files in three formats; Roary-CSV, CSV and Rtabstruct_presence_absence.Rtab: structural variant presnce/absence matrixsummary_statistics.txt: summary of gene frequencies based on RoaryVCF: directory containing VCF files for each cluster generated by SNP-SITESaligned_gene_sequences: directory of alignment files for each cluster in FASTA formatGFF: directory of GFF files for each sample in GFF3 formatggc_data: intermediate datastructures written to disk, required for querying.
Querying the graph#
We can now query the graph. To do so, run:
ggcaller --query CPS_queries.fasta --graph Bentley_et_al_2006_CPS_sequences/input.gfa --colours Bentley_et_al_2006_CPS_sequences/input.color.bfg --data ggc_Bentley_et_al_CPS/ggc_data --out ggc_Bentley_et_al_CPS --threads 4
Results will be saved in ggc_Bentley_et_al_CPS/matched_queries.fasta.
Details on the output can be found in Interpreting results.
From matched_queries.fasta, we can see that all the genes queried were identified in the graph.
As we searched for specific gene variants, this search was too stringent to return orthologues in other genomes.
Important
We recommend searching for partial gene sequences,
or lowering --query-id to return more distantly related sequences.